BioPathBot
Description
Depuis une databiographie (correction(s): ) suivant le format utilisé sur le wiki (correction(s): kiki, tiki ) (ex (correction(s): exo, en, eu, eux, es, eh, et, x, e, rex, tex ): 1890.03.19 / Genève), le BioPathBot est capable d'extraire les informations temporelles et spatiales. Ces deux données forment un tuple (correction(s): tuile, tulle ) et sont parsées (correction(s): parées, pansées, parlées, passées ) de façon suivante :
- La date est formée d'une année, d'un mois et d'un jour. Si des informations venaient à manquer, la date est complétée par défaut avec le mois de janvier et le premier du mois.
- Le lieu est transformé en coordonnées géographiques (utilisant la librairie python [https (correction(s):
)://pypi.python.org (correction(s): orge, ore, ors, or, ort, ord, erg )/pypi/geopy (correction(s): ) geopy (correction(s): )])
Ainsi transformées et triées chronologiquement, ces données permettent de générer une carte utilisant [http (correction(s): )://matplotlib (correction(s): ).org (correction(s): orge, ore, ors, or, ort, ord, erg )/basemap (correction(s): )/index.html basemap (correction(s): )], où sont dessinés des points pour marquer les lieux, ainsi que des traits mettant en avant la trajectoire de la personne. Cette carte est ensuite importée sur wikipast (correction(s): ) puis insérée sur une page annexe.
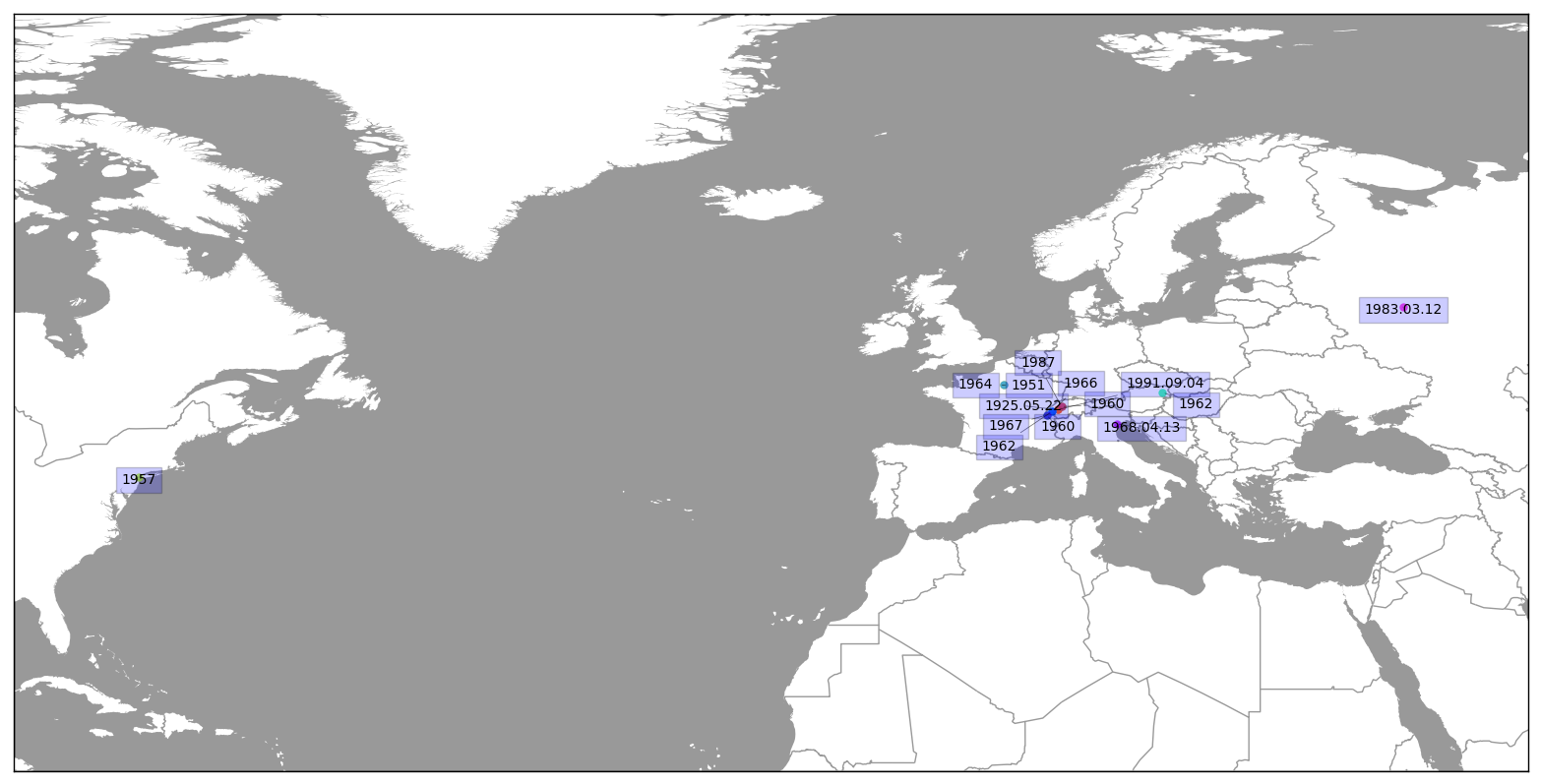
Par exemple la carte de Jean Tinguely se visualise comme:
Critique
- Peu importe le type de page (une page concernant une personne, un hypermot...), une carte est générée. Donc si une page ne relate pas la vie d'une personne, la carte aura peu de sens.
- Avec l'algorithme actuel, les dates et les lieux sont parfaitement extraits et placé sur la carte. Cependant, si les données ne sont pas suffisamment précises, elles seront approximées (correction(s):
) (1980 devient 1980.01.01 tandis (correction(s): candis, tendis, taudis ) que le point géographique correspondant à la Suisse sera son centre géographique). Cela peut mener à des erreurs de trajectoire lors du tri chronologique ou simplement à une position différente de la réalité sur la carte.
- S'il existe des événements post-mortem (correction(s): mortel, mortes, morte
) mais que la mort de la personne n'est pas mentionnée ou ne suit pas la convention de l'hypermot Décès, ces données seront aussi extraites et placées sur la carte, alors même que la personne ne se déplace pas.
- Si des événements sont spatialement trop proches, ils vont se superposer sur la carte et il deviendra difficile de retracer les événements d'une façon visuellement claire. Par exemple, pour un trait entre Berne et Lausanne, il sera difficile de juger si une personne a fait plusieurs fois ce même déplacement ou ne l'a fait qu'une seule fois au cours de sa vie.
- Parfois l'outil qui transforme les lieux en coordonnées géographiques crée des erreurs. Par exemple "Rome" devient "Lomé" qui se situe au Togo.
- Certains événements se passe sans que la personne soit physiquement présente, mais on ne peut pas le détecter automatiquement. Les lieux apparaissent donc sur la carte.
Code
import urllib.request import requests from bs4 import BeautifulSoup import re import math import numpy as np import datetime import random import copy from geopy.geocoders import Nominatim from mpl_toolkits.basemap import Basemap import matplotlib.pyplot as plt from colorsys import hsv_to_rgb from matplotlib.colors import rgb2hex import pdb import time import itertools from geopy.exc import GeocoderTimedOut SEGMENTS = 100 # draw plots inline rather than in a seperate window # %matplotlib inline # draw plots bigger plt.rcParams["figure.figsize"] = [20.0, 10.0] bot_user='BioPathBot' passw='chkiroju' baseurl='http://wikipast.epfl.ch/wikipast/' summary='Wikipastbot update' protected_logins=["Frederickaplan","Maud","Vbuntinx","Testbot","IB","SourceBot","PageUpdaterBot","Orthobot","BioPathBot","ChronoBOT","Amonbaro","AntoineL","AntoniasBanderos","Arnau","Arnaudpannatier","Aureliver","Brunowicht","Burgerpop","Cedricviaccoz","Christophe","Claudioloureiro","Ghislain","Gregoire3245","Hirtg","Houssm","Icebaker","JenniCin","JiggyQ","JulienB","Kl","Kperrard","Leandro Kieliger","Marcus","Martin","MatteoGiorla","Mireille","Mj2905","Musluoglucem","Nacho","Nameless","Nawel","O'showa","PA","Qantik","QuentinB","Raphael.barman","Roblan11","Romain Fournier","Sbaaa","Snus","Sonia","Tboyer","Thierry","Titi","Vlaedr","Wanda"] depuis_date='2017-02-02T16:00:00Z' liste_pages=[] for user in protected_logins: result=requests.post(baseurl+'api.php?action=query&list=usercontribs&ucuser='+user+'&format=xml&ucend='+depuis_date+'&ucshow=new') soup=BeautifulSoup(result.content,'lxml') for primitive in soup.usercontribs.findAll('item'): title = primitive['title'] if 'Fichier' not in title and 'BioPathBot' not in title: liste_pages.append(primitive['title']) names=list(set(liste_pages)) for title in names: print(title) # Login request payload={'action':'query','format':'json','utf8':,'meta':'tokens','type':'login'} r1=requests.post(baseurl + 'api.php', data=payload) #login confirm login_token=r1.json()['query']['tokens']['logintoken'] payload={'action':'login','format':'json','utf8':,'lgname':bot_user,'lgpassword':passw,'lgtoken':login_token} r2=requests.post(baseurl + 'api.php', data=payload, cookies=r1.cookies) #get edit token2 params3='?format=json&action=query&meta=tokens&continue=' r3=requests.get(baseurl + 'api.php' + params3, cookies=r2.cookies) edit_token=r3.json()['query']['tokens']['csrftoken'] edit_cookie=r2.cookies.copy() edit_cookie.update(r3.cookies) #setup geolocator geolocator = Nominatim(timeout=30) # upload config def uploadMap(filename): # read local file upload_file = open(filename,"rb") upload_contents = upload_file.read() upload_file.close() # setting parameters for upload # ref: https://www.mediawiki.org/wiki/API:Upload payload={'action':'upload','filename':filename, 'ignorewarnings':1, 'token':edit_token} files={'file':upload_contents} # upload the image print("Uploading file to %s via API..." % (baseurl+"index.php/Fichier:"+filename)) r4=requests.post(baseurl+'api.php',data=payload,files=files,cookies=edit_cookie) # in case of error print the response # print(r4.text) # add link to biopath in original page if not already existing def addLinkToOriginalPage(name): result=requests.post(baseurl+'api.php?action=query&titles='+name+'&export&exportnowrap') soup=BeautifulSoup(result.text, "lxml") #soup=BeautifulSoup(result.text) code= for primitive in soup.findAll("text"): code+=primitive.string exist = re.findall("(\[\["+name+" BioPathBot\]\])",code) if(len(exist)==0): title = name content = "\n\n"+""+name+" BioPathBot" requests.post(baseurl+'api.php?action=query&titles='+title+'&export&exportnowrap') payload={'action':'edit','assert':'user','format':'json','utf8':,'appendtext':content,'summary':summary,'title':title,'token':edit_token} r4=requests.post(baseurl+'api.php',data=payload,cookies=edit_cookie) def addToPage(name, images, legend): title = name + " BioPathBot"
content = ""+name+"
"+'
'
for img in images:
content += "
"
pageToChange = requests.post(baseurl+'api.php?action=query&titles='+title+'&export&exportnowrap')
payload={'action':'edit','assert':'user','format':'json','utf8':,'text':content,'summary':summary,'title':title,'token':edit_token}
r4=requests.post(baseurl+'api.php',data=payload,cookies=edit_cookie)
print(r4.text)
# BioPathBot : add line of databiographie to the right page (time and space)
def getDataFromPage(name):
data = []
dates = []
places = []
print("Page Created: " + name)
result=requests.post(baseurl+'api.php?action=query&titles='+name+'&export&exportnowrap')
soup=BeautifulSoup(result.text, "lxml")
#soup=BeautifulSoup(result.text)
code=
for primitive in soup.findAll("text"):
if primitive.string:
code+=primitive.string
# split on list (*)
lines = code.split("*")
for line in lines :
# add breaking lines (otherwise will be appened directly in one line)
line = "\n\n"+line
# get date if exist
date = re.findall("((?<=\[\[)\d*(\.*\d*\.*\d*)*(?=\]\]))",line)
dateToAdd = ""
if len(date) != 0 :
dateToAdd = date[0][0]
# get place if exist
place = re.findall("(?<=\/\s\[\[)[A-zÀ-ÿ\s\-]*(?=\]\])",line)
if(len(place)==0):
place = re.findall("(?<=\/\[\[)[A-zÀ-ÿ\s\-]*(?=\]\])",line)
placeToAdd = ""
if len(place) != 0:
placeToAdd = place[0]
if placeToAdd == "Rome":
placeToAdd = "Roma"
# if both the date and the location are available, append in data array
if dateToAdd and placeToAdd:
location = ""
for retries in range(5):
try:
location = geolocator.geocode(placeToAdd)
except GeocoderTimedOut:
continue
break
# geopy usage policy max 1 request/sec
# https://operations.osmfoundation.org/policies/nominatim/
time.sleep(2)
if location:
print("Location: " + placeToAdd + " : " + str(location.longitude) + "," + str(location.latitude))
dataToAdd = [location.longitude,location.latitude];
dates.append(dateToAdd)
places.append(placeToAdd)
data.append(dataToAdd)
# stop getting data if find Décès
foundDeces = re.findall("(\[\[Décès*\]\] (de |d)\[\["+name+")",line)
if(len(foundDeces) != 0):
break
return [data, dates, places]
# finds the minimal and maximal longitude and latitude
def findCorners(pts):
minlon = maxlon = pts[0][0]
minlat = maxlat = pts[0][1]
for p in pts:
currlon = p[0]
if currlon<minlon:
minlon = currlon
elif currlon>maxlon:
maxlon = currlon
currlat = p[1]
if currlat<minlat:
minlat = currlat
elif currlat>maxlat:
maxlat = currlat
return [minlon, maxlon, minlat, maxlat]
# draws the map, some points and the lines
def drawmap_colors(pts, dates, places, filename, export=False):
n_pts = len(pts)
corners = findCorners(pts)
txt = ""
m = Basemap(llcrnrlon=corners[0]-1, llcrnrlat=corners[2]-1, urcrnrlon=corners[1]+1, urcrnrlat=corners[3]+1, resolution='i')
m.drawmapboundary(fill_color='0.6')
m.drawcountries(linewidth=1.0, color='0.6')
m.fillcontinents(color='white', lake_color='white')
for i in range(n_pts-1): # draw lines
for j in range(SEGMENTS):
start = pts[i] + (pts[i+1]-pts[i])*(j/SEGMENTS)
end = pts[i] + (pts[i+1]-pts[i])*((j+1)/SEGMENTS)
m.plot([start[0], end[0]], [start[1], end[1]], color=hsv_to_rgb((i+j/SEGMENTS)/n_pts, 1, 1))
for i in range(n_pts): # draw points
curr_color = hsv_to_rgb(i/n_pts, 1, 1)
m.plot(pts[i][0], pts[i][1], marker='o', color=curr_color, fillstyle='full', markeredgewidth=0.0)
txt += "
" + dates[i] + " / " + places[i] + ". "
if export:
plt.savefig(filename, bbox_inches='tight')
plt.close()
# plt.show()
return txt
# inp: point inside the box
# hs: half dimensions of the box
# outp: another point
# finds the intersection of the segment inp-outp and the box
def line_box(inp, hs, outp):
dir = outp - inp
if dir[0] == 0: dir[0] = np.nextafter(0, 1)
if dir[1] == 0: dir[1] = np.nextafter(0, 1)
ref = np.array([np.copysign(hs[0],dir[0]), np.copysign(hs[1],dir[1])])
dir_x = np.array([(ref[1]/dir[1])*dir[0], ref[1]])
dir_y = np.array([ref[0], (ref[0]/dir[0])*dir[1]])
fdir = dir_x if np.linalg.norm(dir_x) < np.linalg.norm(dir_y) else dir_y
return inp+fdir
# computes the repulsion force if 2 boxes are overlapping
def repulsion_force(pos1, pos2, bbox1, bbox2):
hs1 = np.array([bbox1.width,bbox1.height])/2
hs2 = np.array([bbox2.width,bbox2.height])/2
c1 = pos1 + hs1
c2 = pos2 + hs2
# if both same position, choose a random direction
if all(c1==c2):
return (np.random.rand(2)-np.array([0.5,0.5]))*hs1[1]
b1 = line_box(c1, hs1*1.5, c2)
b2 = line_box(c2, hs2*1.5, c1)
# if pointing in the opposite direction
if np.dot(b1-c1,b2-b1) < 0:
return b2-b1
return np.zeros(2)
# readjust text labels so there is no overlap
def adjust_text(texts, text_width, text_height,num_iterations=20,eta=0.5):
text_pos = [np.array(text.get_position()) for text in texts]
indices = list(range(len(texts)))
colliding = [True for text in texts]
# get text bounding boxes
f = plt.gcf()
r = f.canvas.get_renderer()
ax = plt.gca()
bboxes = [text.get_window_extent(renderer=r).transformed(ax.transData.inverted()) for text in texts]
# center text pos on markers
for i in range(len(texts)):
text_pos[i][0] -= bboxes[i].width/2
text_pos[i][1] -= bboxes[i].height/2
# readjust text labels
for _ in range(num_iterations):
random.shuffle(indices)
for (i,j) in itertools.combinations(indices, 2):
if i == j:
continue
# pdb.set_trace()
f = repulsion_force(text_pos[i], text_pos[j], bboxes[i], bboxes[j])
text_pos[i] += f*eta
# delete text objects and create annotations on the readjusted positions
for i in range(len(texts)):
a = plt.annotate(texts[i].get_text(), xy=texts[i].get_position(), xytext=text_pos[i],
arrowprops=dict(arrowstyle="-", color='k', lw=0.5, alpha=0.6),bbox=dict(facecolor='b', alpha=0.2))
# plt.plot(text_pos[i][0], text_pos[i][1], marker='o',color='r', fillstyle='full', markeredgewidth=0.0,alpha=0.7)
# plt.plot(text_pos[i][0]+bboxes[i].width, text_pos[i][1], marker='o',color='r', fillstyle='full', markeredgewidth=0.0,alpha=0.7)
# plt.plot(text_pos[i][0]+bboxes[i].width, text_pos[i][1]+bboxes[i].height, marker='o',color='r', fillstyle='full', markeredgewidth=0.0,alpha=0.7)
# plt.plot(text_pos[i][0], text_pos[i][1]+bboxes[i].height, marker='o',color='r', fillstyle='full', markeredgewidth=0.0,alpha=0.7)
a.draggable();
texts[i].remove()
# draws the map, some points and the lines
def drawmap_date(pts, dates, places, filename, export=False):
n_pts = len(pts)
corners = findCorners(pts)
txt = ""
# ratio correction to 2:1
lon_width = min((corners[1]-corners[0]+10)*1.1, 360)
lat_width = min((corners[3]-corners[2]+10)*1.1, 180)
lon_center = (corners[1]+corners[0])/2
lat_center = (corners[3]+corners[2])/2
if lon_width > lat_width*2:
lat_width = lon_width/2
else:
lon_width = lat_width*2
corners[0] = lon_center-lon_width/2
corners[1] = lon_center+lon_width/2
corners[2] = lat_center-lat_width/2
corners[3] = lat_center+lat_width/2
if corners[0] < -180:
corners[1] -= corners[0]-(-180)
corners[0] = -180
elif corners[1] > 180:
corners[0] -= corners[1]-180
corners[1] = 180
if corners[2] < -90:
corners[3] -= corners[2]-(-90)
corners[2] = -90
elif corners[3] > 90:
corners[2] -= corners[3]-90
corners[3] = 90
# draw map background
m = Basemap(llcrnrlon=corners[0], llcrnrlat=corners[2], urcrnrlon=corners[1], urcrnrlat=corners[3], resolution='i')
m.drawmapboundary(fill_color='0.6')
m.drawcountries(linewidth=1.0, color='0.6')
m.fillcontinents(color='white', lake_color='white')
texts = []
for i in range(n_pts-1): # draw lines
for j in range(SEGMENTS):
start = pts[i] + (pts[i+1]-pts[i])*(j/SEGMENTS)
end = pts[i] + (pts[i+1]-pts[i])*((j+1)/SEGMENTS)
m.plot([start[0], end[0]], [start[1], end[1]], color=hsv_to_rgb((i+j/SEGMENTS)/n_pts, 1, 1))
for i in range(n_pts): # draw points
curr_color = hsv_to_rgb(i/n_pts, 1, 1)
x,y = m(pts[i][0], pts[i][1])
m.plot(x, y, marker='o', color=curr_color, fillstyle='full', markeredgewidth=0.0,alpha=0.7)
texts.append(plt.text(x, y, dates[i]))
txt += "" + dates[i] + " / " + places[i] + ".
"
adjust_text(texts, 1, 0.4)
if export:
plt.savefig(filename, bbox_inches='tight')
plt.close()
return txt
for name in names:
image_filename_colors = (name + "_colors_biopath.png").replace(" ","_")
image_filename_date = (name + "_date_biopath.png").replace(" ","_")
data = getDataFromPage(name)
if len(data[0]) != 0:
legend_colors = drawmap_colors(np.array(data[0]), data[1], data[2], image_filename_colors, True)
drawmap_date(np.array(data[0]), data[1], data[2], image_filename_date, True)
uploadMap(image_filename_date)
uploadMap(image_filename_colors)
addToPage(name, [image_filename_colors, image_filename_date], legend_colors)
addLinkToOriginalPage(name)
print("end")
Groupe
| Nom et Prénom | Pseudo |
|---|---|
| Christophe Badoux | Christophe |
| Julien Burkhard | JulienB |
| Kim Lan Phan Hoang | Kl |
| Robin Lang | Roblan11 |